The point: Not a real post, just some happy news.
A few months ago, I took down this blog, as I'd become a little embarrassed about it. I started this blog on VASP and DFT when I first started graduate school with no experience in DFT calculations and minimal programming abilities. It's scary to know that anyone on the internet can see how much of a beginner I was (and still am). Even now, after coming a long way and learning a lot more, I'm primarily an experimentalist that only dabbles in DFT...
Anway, I decided to bring the blgo back because it does seem like it's still helpful to people. I was very surprised to find that my blog had been cited in a publication! Wow!
Yes... it's from last year, but I only saw it last week....
I usually try to keep these notes short and straight to the point, but I wanted to take some time to say that I'm pretty happy if these notes are useful to others. I apologize that I don't update frequently and that sometimes my notes aren't the best. I hope to someday clean things up so that they're something that I'm more proud of. Even so, thanks for reading.
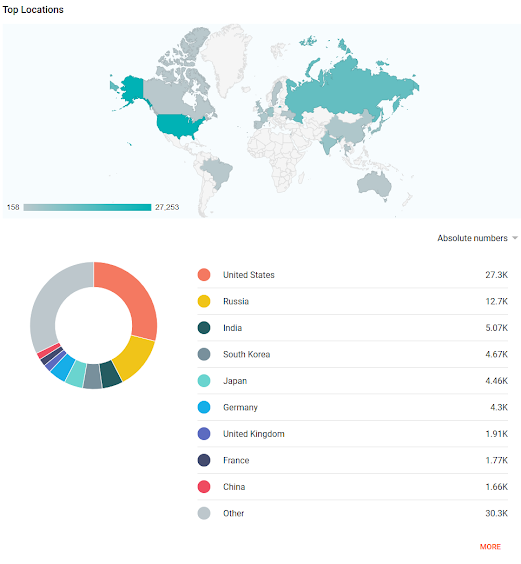
I'll leave off with some interesting stats snapshots:
Cheers!